Machine-Intelligence, Computing & Security Covid-19 Initiatives
- A Controlled Response to COVID-19
- DeepGLEAM COVID-19 Forecast Now Part of CDC Model
- Distributed Control and Hierarchical Decision Making for Ventilated Patients
- High-Throughput Wastewater SARS-CoV-2 Detection Enables Forecasting of Community Infection Dynamics in San Diego County
- Privacy-preserving COVID-19 Discovery
- Real-time Phylogenetic Inference and Transmission Cluster Analysis of COVID-19
- Robust AI for Automated and Accelerated Literature and Trend COVID-19 Systemization of Knowledge
- Technical University Darmstadt & UC San Diego Privacy-preserving Contact Tracing App
- Ultrafast Sample placement on Existing tRees (UShER), Real-time Virus Genomic Surveillance and Contact Tracing
- UV-Drone: Mobile Disinfection Platform for Community Facilities with Minimum Human Exposure
A Controlled Response to COVID-19
Lead by Professor Massimo Franceschetti and Professor Behrouz Touri
We are proposing a generalization of Susceptible Infectious Recovered (SIR) model for inhomogeneous population. Using tools from optimal control and nonlinear control and based on the proposed mean-field model, we find the optimal-cost containment policy to meet the local health-care provider's care capacity.
UPDATE April 2021
Abstract: In this research project, theoretically, we show that for a large population, a simple (and easy-to-analyze) macro-level dynamics closely approximates a micro-level and complex model of COVID-19 infection in a population. Further, we have developed a system identification (parameter estimation) method to estimate the parameters of the (easy to analyze) obtained model which leads to a prediction method for infection as well as mortality of the pandemics in a given society. Our results also enable us to synthesis and study optimal control strategies for COVID (and possibly other pandemics) in the future. Currently, we are working on an automatic method for detecting phase transitions in the pandemics and we are showing the efficacy of our developed methods on epidemics propagation for several regions (including Japan and California).
Back to Top
DeepGLEAM COVID-19 Forecast Now Part of CDC Model
Lead by Professor Rose Yu and Yian Ma
UPDATE April 2021
A computational model that forecasts the number of COVID-19 deaths in the United States as a whole and in each state, which was developed by a team of researchers from the University of California San Diego and Northeastern University, is now part of the national mortality forecast issued by the Centers for Disease Control. The UC San Diego-Northeastern model, called DeepGLEAM, is unique because it combines a physics-based model, known as GLEAM with deep learning, a computing system made up of algorithms inspired by the way the human brain is organized. The hybrid model leverages rich data information about COVID 19 from the real world--for example when a person had been infected and where they have traveled. “Combining the two is important,” said Yu, the computer scientist. The physics-based model is not good at handling uncertainty and unknowns in the data, for example how well people adhere to travel bans. That’s where deep learning comes in to help reduce uncertainty. For full article click here.
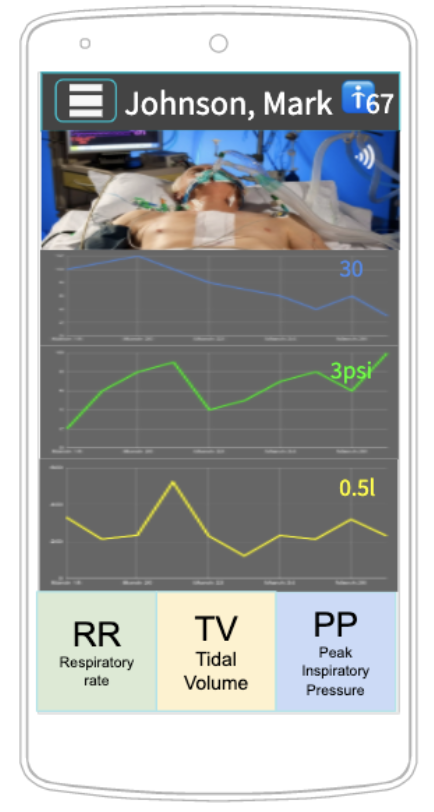
Distributed Control and Hierarchical Decision Making for Ventilated Patients
Lead by PhD Student Michael Barrow
Michael Barrow in close collaboration with Ryan Kastner and Dr. Shanglei Liu (former UC San Diego resident, now fellow at the Mayo Clinic) and their team of nearly 15 are addressing the pending need to scale out the care of ventilated patients. Right now, major efforts are underway to increase the ventilator supply -- a critical task and one that is likely to result in a number of different solutions. Once ventilated, patients require care from trained experts like doctors and respiratory therapists. Unfortunately, we can quickly reach the point where the number of ventilated patients outnumbers those that can monitor them. We are developing a low-cost telemedicine system that allows clinicians to simultaneously manage a large number of patients on ventilators. We anticipate that this will help scale the decision-making ability of clinical experts enabling them to more efficiently monitor and care for a large number of ventilated patients. To learn more click here.
UPDATE April 2021
One of our main goals is to improve the efficiency of medical treatment. Nowadays, hardware is no more the limiting factor but medical expertise is. Every time a doctor enters a room with a sick patient, he or she needs to put on a bunch of medical equipment like a PPE, a face mask, gloves, a face shield, etc. All this just to make a simple decision and look at some graphs. Our telemedicine app and the accompanying ventilator will enable doctors to look at such graphs and make such decisions remotely. This will save a ton of time and PPE usage, and the doctors will be able to help more patients!
At the end of this quarter, we plan on testing our ventilator on a mechanical lung with our collaborators in the pulmonary department. To view our team update presentation click here.
High-Throughput Wastewater SARS-CoV-2 Detection Enables Forecasting of Community Infection Dynamics in San Diego County
Lead by Tara Javidi, Rob Knight, Smruthi Karthikeyan, Nancy Ronquillo, Pedro Belda-Ferre, Destiny Alvarado, Christopher A. Longhurst
Developed in a first collaboration between MICS and the Center for Microbiome Innovation, the wastewater sampling initiative began on our campus in the past summer, but its expansion was bottlenecked by the manual effort required to concentrate samples. This issue was overcame in a first-ever joint effort between the Center for Microbiome Innovation (CMI) and the Center for Machine-Intelligence, Computing & Security (MICS) to develop an automated system. This new system removes the previous potential for human error as well as increased the number of samples processed concurrently, allowing for more rapid detection of positive results. Read the full published research paper here.
Back to TopPrivacy-preserving COVID-19 Discovery
Lead by Professor Farinaz Koushanfar
Several research labs worldwide, both on the industrial and university side, are focused on sequencing the COVID-19 virus and studying its interactions in-vivo and in-vitro. In several cases, sharing the data and models is not possible due to privacy and IP issues, impeding collaboration for achieving a faster discovery. We provide one-of-a-kind provable privacy-preserving methodologies, based on verified cryptographic protocols, which enable collaboration among the various entities in the encrypted domain. In such a way, the privacy of the IP/content owners is not compromised, while new collaborations and discoveries are uniquely enabled. In particular, multiple research labs will be able to jointly work on the encrypted version of the aggregated data without disclosing their sensitive information to any other research lab. Our state-of-the-art technologies provide a secure platform such that the confidentiality of the data during computation is guaranteed. The platform also assured the correctness of the computation; the result is equivalent to running the underlying algorithm on the cleartext (unencrypted) data. The platform will remove several critical obstacles in the global-scale study of COVID-19, and in turn, will accelerate the process of finding new recovery mechanisms.
Back to TopReal-Time Phylogenetic Inference and Transmission Cluster Analysis of COVID-19
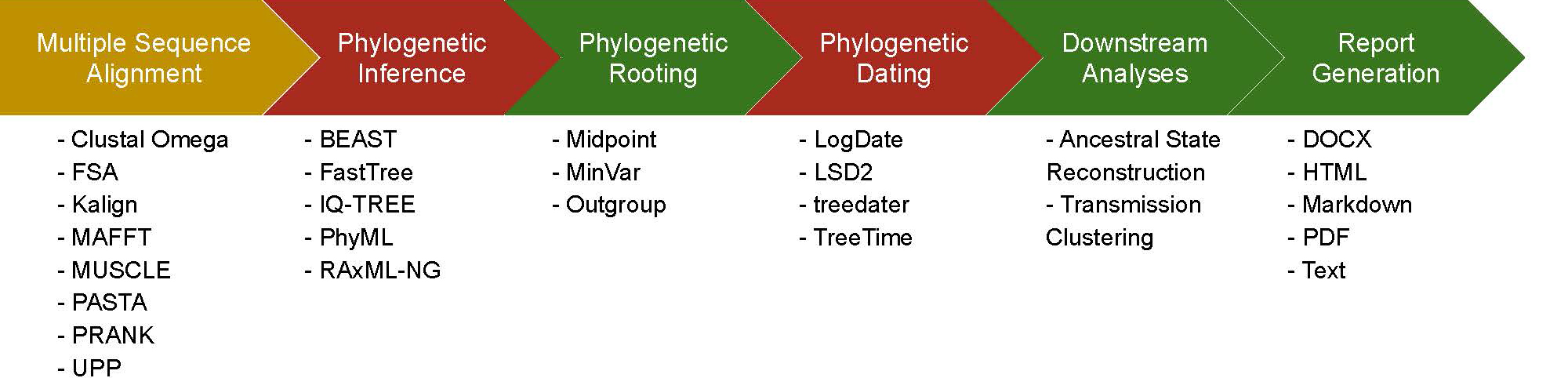
Lead by PI Niema Moshiri and Co-PI Tajana S. Rosing
COVID-19, was first documented in Wuhan, China. Within months, the virus spread rapidly around the world, and despite governmental preventative measure, the global number of cases continues to grow exponentially. In order to fight this growth, public health officials need to be able to answer questions such as "How is the virus spreading through the population?" and "How many individual outbreaks exist within a given community?". Such questions are often investigated using contact tracing, but with limited availability of COVID-19 tests, contact tracing is error-prone. For viral pathogens, isolates from different hosts will likely have genetic differences, and a phylogenetic analysis of these differences can provide critical information for answering the aforementioned public health questions. With the increasing accessibility and affordability of the sequencing, the number of available COVID-19 genomes assemblies is growing rapidly, which in turn creates a need for scalable methods to enable real-time analysis and dissemination. The ability to conduct such analyses in real-time is limited primarily by two key factors: (1) limited computational training by virologists and public health officials to be able to initiate the analyses, and (2) limited access to sufficient high-performance computing resources to be able to execute the analyses. This project's goal is to develop a user-friendly, scalable, and modular workflow for conducting a computational phylogenetic analysis of assembled viral genomes. Our solution includes: (1) the development of a novel software tool for orchestrating the automated end-to-end workflow, (2) the development of novel algorithms (and software implementations of these algorithms) to speed up the computational bottlenecks of the workflow, (3) the development of novel hardware systems for accelerating the workflow, and (4) a real-time publicly-accessible repository in which researchers can access the most up-to-date analysis results (with intermediate files) of all COVID-19 genomes currently available to prevent repeat computation efforts. To learn more click here.
UPDATE April 2021
Much has been accomplished in the course of this year!
- We successfully accelerated the Multiple Sequence Alignment step of a viral phylogenetic analysis pipeline
- A process that used to take hours/days to run on even 100,000 SARS-CoV-2 genomes now runs in around 1 minute
- The speedup widens as the number of genomes increases, and in present day, we're at over 1 million SARS-CoV-2 genomes worldwide
- Open-Source Software Tool (ViralMSA): https://github.com/niemasd/ViralMSA
- ViralMSA was also presented at the following conferences:
- Bioinformatics Community Conference (BCC) 2020
- Computing Research Association (CRA) Virtual Conference 2020
- COVID-19 Dynamics & Evolution Conference
- Cold Spring Harbor Laboratory (CSHL) Biological Data Science Meeting 2020
- We accelerated many bioinformatics algorithms
- We have a number of in-progress results on the other steps of the SARS-CoV-2 sequence analysis pipeline, which will be presented at the upcoming 28th International Dynamics & Evolution of Human Viruses Conference:
- "FPGA-based acceleration of primer trimming"
- "A GPU-Powered Phylogenetic Analysis for Large-scale Genomic Sequences"
- "An evaluation of phylogenetic workflows in viral molecular epidemiology"
- "Optimizing high-performance cloud computing to enable real-time high-throughput SARS-CoV-2 whole genome sequence analysis"
- N. Moshiri, “ViralMSA: massively scalable reference-guided multiple sequence alignment of viral genomes” Bioinformatics. Btaa743. doi:10.1093/bioinformatics/btaa743
- Nominated for the best paper award at a top conference in Dr. Rosings area.
- B. Khaleghi, N. Moshiri, T. Rosing, S. Salamat, S. Shubhi, "FPGA Acceleration of Protein Back-Translation and Alignment" , IEEE/ACM Design Automation and Test in Europe Conference (DATE), 2021.
- Saransh Gupta, Mohsen Imani, Behnam Khaleghi, Niema Moshiri, and Tajana S. Rosing “RAPIDx: A ReRAM Processing in-Memory Architecture for DNA Short Read Alignment”, 70th Virtual Meeting of The American Society of Human Genetics (ASHG), 2020
- Mohsen Imani, Yeseong Kim, Niema Moshiri, Tajana Rosing. “GenieHD: Efficient DNA Pattern Matching Accelerator using Hyperdimensional Computing”, IEEE Design, Automation & Test in Europe Conference & Exhibition (DATE), 2020. (Best paper candidate)
For further details in regards to these publications please see Dr. Moshiri’s "Research & Publications" page.
Back to TopRobust AI for Automated and Accelerated Literature and Trend COVID-19 Systemization of Knowledge
Lead by Professor Farinaz Koushanfar
We are developing novel robust and safe accelerated methodologies, based on natural language processing (NLP) to systemize the knowledge discovery and trends in COVID-19 domain. While several disparate entities are working in research on the topic, and a multitude of sources are publishing news on a daily basis, it is interactive for researchers and public health professionals to follow the vast updates. NLP-based methods are useful for automated knowledge discovery but suffer from the existence of unreliable sources, data poisoning, and fake news. We focus on making the automated knowledge search systematic, safe and robust, while we simultaneously address scalability through new hardware/software/algorithm co-design.
 Back to Top
Back to TopTechnical University Darmstadt & UC San Diego Privacy-preserving COVID-19 Bluetooth Contact Tracing App
Lead by Professor Farinaz Koushanfar

Farinaz Koushanfar has partnered with collaborators in San Diego and at the Technical University Darmstadt in Germany to develop a contact tracing app with user privacy and security as the primary priority. The engineer and machine learning specialist began lending her expertise to the project more than a month ago, as COVID-19 contact tracing efforts raised privacy and security concerns. Koushanfar collaborates with crypto and security researchers, policymakers and colleagues abroad to design a reliable and trustworthy solution in the form of a Bluetooth-based contact tracing app. The app is called TraceCORONA and works in a fully anonymous manner. It transmits encrypted data to a server that stores information in random streams to protect the user’s identity. The app has been released for beta testing on Android and can be downloaded at https://tracecorona.net/download-tracecorona/. An iOS version will follow.
UPDATE April 2021
To satisfy the needs of users and encourage user installation, TraceCORONA has the following intriguing features: (1) Anonymity without compromises using cryptographic Encounter Tokens; (2) Secure messaging with health experts and organizations; (3) Prevention of fake news; (4) Interoperability with other applications over the secure communication channel of TraceCORONA. With the capability of TraceCORONA, users can opt-in to various services, including confidential and trustworthy interactions with healthcare providers, testing centers, health authorities, and hospitals. These services will be implemented using secure messaging, secure document exchange and secure health process capabilities provided by the platform. For our latest developments and news view TU Darmstadt's blog.
Ultrafast Sample placement on Existing tRees (UShER), Real-time Virus Genomic Surveillance and Contact Tracing
Lead by Yatish Turakhia, Russell Corbett-Detic and Angie Hinrichs
UShER identifies the relationships between a user’s newly sequenced viral genomes and all known SARS-CoV-2 virus genomes by adding them to an existing phylogenetic tree, a branching diagram like a family tree that shows how the virus has evolved in different lineages as it accumulates mutations.
“We are able to maintain a comprehensive phylogenetic tree of more than 1.2 million coronavirus sequences and update it with new sequences in real time. No other tool can handle trees of this size with a comparable efficiency,” said first author Yatish Turakhia, a postdoctoral scholar at the Genomics Institute. “This helps us keep track of all variants in circulation, including new variants that are emerging.”
UShER and related data visualization tools are available to the research community through the UCSC SARS-CoV-2 Genome Browser, which also provides access to a wide range of data and results from ongoing scientific research on the virus, including new variants that are especially concerning.
This work has since been featured in articles with Academic Times, Eureka Alert, MedicalXpress, Mercury News, News Medical
(Original Story from UC Santa Cruz and Technology Networks)
Back to Top

UV-Drone: Mobile Disinfection Platform for Community Facilities with Minimum Human Exposure
Lead by Tara Javidi and DetecDrone Team
Disinfecting areas that have been exposed to COVID-19 using conventional method of washing and cleaning of surfaces and spaces with disinfecting liquid/materials, exposes the most disadvantaged workers at the frontline of fighting the disease and increases the risk of exposures and disease contraction for the cleaning crew.
This project leverages Professor Javidi’s existing DetecDrone research platform developed at UC San Diego to significantly improve on the process of UV based disinfection. In addition to providing a cost effective and agile non-contact UV-C sterilization to the UC San Diego Medical facilities, we envision the first Do It Yourself (DIY) prototype of a drone platform for delivering non-contact mobile UV sterilization unit (UVerizeDrone) for community facilities such as schools, institutions of higher education, offices, daycare centers, businesses, and community centers. For the latest information please see UV-C Disinfection Drones project website.
Back to Top





